MOLECULAR BASIS OF INHERITANCE
INTRODUCTION
- Nucleic acids are the building blocks of genetic material.
- Genetic material is that substance which not only controls the formation and expression of traits in an organism but can replicate and pass on from a cell to its daughter cell or from one generation to the next.
- DNA and RNA are two types of genetic material.
EVIDENCES OF DNA AS THE GENETIC MATERIAL
- The following experiments conducted by the molecular biologists provide direct evidences of DNA being the genetic material.
BACTERIAL TRANSFORMATION OR GRIFFITH'S EXPERIMENTS
- Griffith (1928) injected mice with virulent and smooth
(S-type, smooth colony with mucilage) form of Diplococcus pneumoniae. The mice died due to pneumonia. No death occurred when mice were injected with non-virulent or rough (R-type, irregular colony without mucilage) form or
heat- killed virulent form. However, in a combination of heat killed S-type and live R-type bacteria, death occurred in some mice. Autopsy of dead mice showed that they possessed S-type living bacteria, which could have been produced only by transformation of R-type bacteria. - The transforming chemical was found out by O.T.Avery, C.M. Mc. Leod and M. Mc. Carty (1944). They fractionated heat-killed S-type bacteria into DNA, carbohydrate and protein fractions. DNA was divided into two parts, one with DNAase and the other without it. Each component was added to different cultures of R-type bacteria.
- Transformation was found only in that culture which was provided with intact DNA of S-type. Therefore, the trait of virulence is present in DNA. Transformation involves transfer of a part of DNA from surrounding medium or dead bacteria (donor) to living bacteria (recipient) to form a recombinant.
- Transformation was the first step in the identification of genetic material.
EVIDENCE FROM GENETIC RECOMBINATION IN BACTERIA OR BACTERIAL CONJUGATION
- Lederberg and Tatum (1946) discovered the genetic recombination in bacteria from two different strains (mutant strain A and mutant strain B) through the process of conjugation.
- Bacterium Escherichia coli can grow in minimal culture medium containing minerals and sugar only. It can synthesize all the necessary vitamins from these raw materials. But its two mutant strains were found to lack the ability to synthesize some of the vitamins necessary for growth. These could not grow in the minimal medium till the particular vitamins were not supplied in the culture medium.
- Mutant strain A : It (used as male strain) has the genetic composition of Met– , Bio–, Thr+, Leu+, Thi+. It lacks the ability to manufacture vitamins, methionine and biotin and can grow only in a culture medium which contains these vitamins in addition to sugar and minerals.
- Mutant strain B : It (used as female strain or recipient) has a genetic composition Me++, Bio+, Thr–, Leu–, Thi–. It lacks the ability to manufacture threonine, leucine and thionine and can grow only when these vitamins are added to the growing medium.
- These two strains of E.coli are, therefore, unable to grow in minimal culture medium, when grown separately. But when a mixture of these two strains was allowed to grow in the same medium, a number of colonies were formed. This indicates that the portion of donor DNA containing information to manufacture threonine, leucine and thionine had been transferred and incorporated in the recipient's genotype during conjugation.
- This experiment of Lederberg and Tatum showed that the conjugation results in the transfer of genetic material DNA from one bacterium to other. During conjugation, a cytoplasmic bridge is formed between two conjugating bacteria.
EVIDENCE FROM BACTERIOPHAGE INFECTION
- Hershey and Chase (1952) discovered that DNA is the genetic material of bacteriophage.
They conducted their experiment on T2 bacteriophage, which attacks on E.coli bacterium.
- The phage particles were prepared by using radioisotopes of S35 and P32 in the following steps-
- Few bacteriophages were grown in bacteria containing 35S which was incorporated into cysteine and methionine amino acids of proteins and thus, these amino acids with 35S formed the proteins of phage.
- Some other bacteriophages were grown in bacteria having 32P, which was restricted to DNA of phage particles.
- These two radioactive phage preparations (one with radioactive proteins and another with radioactive DNA) were allowed to infect the culture of E.coli. The protein coats were separated from the bacterial cell walls by shaking and centrifugation.
- The heavier infected bacterial cells during centrifugation pelleted to bottom. The supernatant had the lighter phage particles and other components that failed to infect bacteria. It was observed that bacteriophages with radioactive DNA gave rise to radioactive pellets with 32P in DNA. However, in the phage particles with radioactive protein (with 35S) the bacterial pellets have almost nil radioactivity indicating that proteins have failed to migrate into bacterial cell. So, it can be safely concluded that during infection by bacteriophage T2, it was DNA, which entered the bacteria. It was followed by an eclipse period during which phage DNA replicates numerous times within the bacterial cell. Towards the end of eclipse period, phage DNA directs the production of protein coats assembly of newly formed phage particles.
- The above experiment clearly suggests that it is phage DNA and not protein, which contains the genetic information for the production of new bacteriophages. However, in some plant viruses (like TMV), RNA acts as hereditary material (being DNA absent).
Fig. : The Harshey Chase experiment
STRUCTURE OF DNA (DEOXYRIBONUCLEIC ACID)
- The term DNA was given by Zacharis, which is found in the cells of all living organisms except plant viruses,where RNA forms the genetic material and DNA is absent.
- In bacteriophages and viruses, there is a single molecule of DNA, which remains coiled and is enclosed in the protein coat.
- In bacteria, mitochondria, plastids and other prokaryotes, DNA is circular and lies naked in the cytoplasm but in eukaryotes, it is found in the nucleus and is known as the carrier of genetic information and capable of self replication.
- Isolation and purification of specific DNA segment from a living organism was achieved by Nirenberg.
- The chemical analysis has shown that DNA is composed of three different types of compound - sugar molecules, phosphoric acid and nitrogeneous base.
Fig. : DNA
- Sugar molecule : Levene identified a five carbon sugar, ribose in nucleic acid in 1910. It is represented by a pentose sugar, the deoxyribose or 2-deoxyribose which is derived from ribose due to the deletion of oxygen from the second carbon.
- Phosphoric acid : H3PO4 makes DNA acidic in nature.
- Nitrogeneous base : Kossel demonstrated the presence of two pyrimidines (cytosine and thymine) and two purines (adenine and guanine) in DNA and was awarded Nobel Prize in 1910. These are nitrogen containing ring compounds. These are classified into two groups:-
- Purines : Two ring compounds namely as adenine and guanine.
- Pyrimidines : One ring compounds cytosine and thymine. In RNA, uracil is present instead of thymine.
- Nucleosides are formed by a purine or pyrimidine nitrogenous base and pentose sugar. DNA nucleosides are known as deoxyribose nucleosides.
- Nucleotides : In a nucleotide, purine or pyrimidine nitrogenous base is joined by deoxyribose pentose sugar (D), which is further linked with phosphate (P) group to form nucleotides.
- DNA is found in nucleus and cytoplasm. The contribution of cytoplasmic DNA in the cell's total DNA is about 1-5%.
- Rosalind Franklin, studied the structure of DNA using
X-rays. It showed that DNA is a helix. - In 1953, James Watson and Francis Crick proposed the three-dimensional structure of DNA based on X-ray diffraction photographs of DNA fibres taken by Rosalind Franklin and M.H.F. Wilkins.
- For discovering the structure of DNA, Nobel Prize was awarded to Watson, Crick and Wilkins in the year 1962.
- The Watson and Crick model shows that DNA is a double helix with sugar-phosphate backbones on the outside and paired bases on the inside.
- The diameter of DNA molecule is 20 Å.
- The pitch (a complete turn) of DNA has a length of about 34 Å.
- Adjacent bases are separated by 3.4 Å along the helix axis and related by a rotation of 36 degrees.
- There are about 10 base pairs in each turn of DNA double helix.
- The two chains are held together by hydrogen bonds between pairs of bases which helps to stabilize the interaction.
- Hydrogen bonds join the nitrogen bases of one strand with that of the other.
- Adenine - thymine pair has 2 hydrogen bonds while guanine-cytosine pair has 3 hydrogen bonds.
- Adenine always pairs with thymine in DNA and with uracil in RNA. Whereas, cytosine always pairs with guanine in both DNA and RNA.
- Each DNA strand has a backbone of alternate deoxyribose and phosphoric acid groups.
- DNA molecule shows polarity in direction. One end of each chain is called 5' end and the other is called 3' end.
- The sugar-phosphate-sugar component which are joined by phosphodiester bond forms the backbone of DNA duplex.
- The nitrogenous base molecules are attached with the deoxyribose sugar molecules by glycosidic bonds.
- The carbon atoms of the pentose sugar involved in phosphodiester bond formation in DNA are C3 and C5.
- The two strands of DNA (called DNA duplex) are antiparallel and complementary i.e. one in 5' → 3' direction and the other in 3' → 5' direction.
- DNA duplex is made up of two molecules out of which the one that directs the synthesis of the RNA via complementary base pairing is called template or sense strand and the other one is called antisense strand.
- Chemical studies on the bases of DNA were performed by Erwin Chargaff in 1950.
- Pyrimidine bases of DNA are represented by thymine and cytosine and purine bases by adenine and guanine.
- Erwin Charagaff (1950) found that purine and pyrimidine content of DNA are equal.
- According to Chargaff, the percentage of adenine (A) is equal to the percentage of thymine (t) and the percentage of guanine (G) is equal to the percentage of cytosine (C).
- The percentage of A + G equals 50% and the percentage of T + C equals 50%. These relationships are called Chargaffs rules.
- Quantitatively A = T and C = G or
- Chargaff rule also states that A + G = T + C or A + C = G + T.
- While, the base ratio may vary from species to species but it is constant for a given species.
- Base ratio is high in advanced organism and low in primitive organism.
TYPES OF DNAs
- Under different conditions of isolation, purification and crystallization, several forms of DNA have been recognised. They are A-, B-, C- D- and Z-DNA.
- A-, B-, C- and D-DNA are right-handed helices whereas Z-DNA is left-handed helix.
- B-DNA is the most common DNA and are metabolically stable.
- It is believed that during transcription the B-DNA changes to A form.
- The part of the genome in which repetitive sequence are arranged end to end as long tandem arrays is called satellite DNA.
- Satellite DNA was the first type to be identified in the human genome.
- Repetitive DNA occurs in telomere, centromere and ends of transposons.
- In Drosophila, about 25% and in human beings about 30% DNA are repetitive.
- Palindromic DNA is a part of DNA in which the base sequences of one strand is opposite to that of the other strand when read from opposite directions. DNA regions that transcribe rRNA are often palindromic. The true significance of palindromic DNA is, however, not clear, although several function have been suggested like -
- Short palindromes may function as recognition sites of DNA for proteins which also have a two fold rotational symmetry, e.g. lac repressor protein, CRP proteins and many bacterial restriction enzymes.
- Also, gives structural strength to the transcribed RNA by hydrogen bonding in hairpin loops.
- Long palindromic DNA molecules from some lower eukaryotes have been shown to contain genes coding for ribosomal RNA.
3' – C – C – G – G – A – A – T – T – C – C – G – G – 5'
5' – G – G – C – C – T – T – A – A – G – G – C – C – 3'.
- Promiscuous DNA is a special type of DNA which makes movement between mitochondria, chloroplast and nucleus. It was discovered in 1983 in Cambridge University in maize. It was later reported in yeast, mungbean, spinach and peas.
- Repetitive DNA : Multiple copies of DNA having the same or almost the same base pair sequence are constitute repetitive DNA. In higher organisms, 20% - 90% DNA is of this type.
DNA PACKAGING
- The basic unit into which the DNA is packed in the chromatin of eukaryotes is nucleosome.
- A nucleosome contains an octamer of proteins consisting of two each copies of histones H2A, H2B, H3 and H4, around which is wrapped two and a half turns of DNA. Histone H1 bends to DNA outside the ball in the linker region. A typical nucleosome contains 200 bp of DNA helix.
- Nucleosomes in chromatin are seen as beads on string structure when viewed under electron microscope.
- The nucleosomal organization provides a chromatin fibre approximately 10 nm in thickness, which gets further condensed to produce a solenoid of 30 nm diameter. This solenoid structure undergoes further coiling to produce a chromatid of 70 nm diameter which can be seen under the light microscope. A nuclear scaffold formed by non-histone proteins holds all the folded loops of chromatin.
Fig. : Nucleosome
STRUCTURE OF RNA (RIBONUCLEIC ACID)
- RNA is a non - hereditary nucleic acid except in some viruses (retroviruses)
- It is a polymer of ribonucleotide and is made up of pentose ribose sugar, phosphoric acid and nitrogenous base (A, U, G, C).
- Franklin Conrat (1957) establishes that RNA is the genetic material in some viruses.
- RNA may be of two types-genetic and non-genetic.
- Genetic RNA carries the genetic message and is capable of self replication. It is called RNA dependent RNA synthesis.
- Non-genetic RNA are of 3 types mRNA, tRNA and rRNA.
- mRNA (messenger RNA) are formed on specific part of DNA as a complimentary copy of one strand of it in the nucleus. It forms a template for protein synthesis.
- The length of the mRNA is more than the length of the proteins synthesized.
- m-RNA is of 2 types - monocistronic and polycistronic.
- The m-RNA in which genetic signal is present for the formation of only one polypeptide chain is called monocistronic.
- The m-RNA in which genetic signal is present for the formation of more than one polypeptide chains is called polycistronic.
- tRNA (transfer RNA) are also known as soluble RNA, supernatant RNA or adapter RNA
- tRNA is a small RNA chain that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis during translation.
In 1965, R.W. Holley proposed a clover leaf model of tRNA. 3-D structure of tRNA was proposed by S.H. Kim in 1973. tRNA has four sites-
Fig. : tRNA - the adapter molecule
- Amino acid attachment site
- Amino acid recognition site (DHU loop)
- Codon recognition site (anticodon loop)
- Ribosome recognition site (GTPCG loop)
- rRNA (ribosomal RNA) is called insoluble RNA whose function is to attract and provide large surface for spreading of mRNA over ribosomes during translocation process of protein synthesis.
DNA REPLICATION
- DNA is the only molecule capable of self duplication so it is termed as a living molecule.
- All living beings have the capacity to reproduce because of DNA.
- DNA replication takes place in S-phase of the cell cycle. At the time of cell division, it divides in equal parts in the daughter cells.
- Delbruck suggested three methods of DNA-replication i.e.
- Dispersive
- Conservative
- Semi-conservative
DISPERSIVE METHOD
During dispersive method, the DNA undergoes fragmentation and then replicatse and joins to form two molecules of DNA.
CONSERVATIVE METHOD
During conservative method, the original DNA molecule is conserved and its copy is synthesized from the medium.
SEMI CONSERVATIVE MODE OF DNA REPLICATION
Semi conservative mode of DNA replication was first theoretically proposed by Watson and Crick. Later on, it was experimentally proved by Meselson and Stahl (1958) on E.coli and Taylor on Vicia faba.
Meselson and Stahl (1958) cultured (Escherichia coli) bacteria in a culture medium containing N15. After these had been replicated for a few generations in the medium, both strands of their DNA contained N15 as constituents of purines and pyrimidines. When these bacteria with N15 were transferred in cultural medium containing N14, it was found that DNA separated from fresh generation of bacteria possesses one strand heavier than the other. The heavier strand represents the parental strand and lighter one is the new one synthesized from the culture indicating semi conservative mode of DNA replication. Circular form of replication is characteristic of prokaryotes.
Fig. : Second generation daughter molecules after DNA replication
- The following steps are included in DNA replication -
Unzipping
- The separation of 2 chains of DNA is termed as unzipping, and it takes place due to the breaking of H bonds.
The process of unzipping starts at a certain specific point which is termed as initiation point or origin of replication. In prokaryotes, there occurs only one origin of replication but in eukaryotes there occur many origin of replication i.e. unzipping starts at many points simultaneously. At the place of origin, the topoisomerase enzyme (a type of endonuclease) induces a cut in one strand of DNA (nicking) to relax the two strands of DNA.
- The enzyme responsible for unzipping (breaking the hydrogen bonds) is helicase (= Swivelase). In the process of unzipping, Mg+2 act as a cofactor. Unzipping takes place in alkaline medium.
- A protein, helix destabilizing protein prevents recoiling of two separated strands during the process of replication.
- An another protein SSB (single stranded DNA binding protein) prevents the formation of bends or loops in separated strands.
DNA-Gyrase, a type of topoisomerase prevents supercoiling of DNA.
Notes: The process of DNA replication takes a few minutes in prokaryotes and a few hours in eukaryotes.
Formation of New Chain
- To start the synthesis of new chain, special type of RNA is required which is termed as RNA Primer.The formation of RNA primer is catalysed by an enzyme - RNA polymerase (primase). Synthesis of RNA-primer takes place in 5' → 3 direction. After the formation of new chain, this RNA is removed.
- For the formation of new chain, nucleotides are obtained from nucleoplasm. In the nucleoplasm, nucleotides are present in the form of triphosphates like dATP, dGTP, dCTP, dTTP etc.During replication, the 2 phosphate groups of all nucleotides are separated. In this process, energy is yielded which is consumed in DNA replication. So, it is clear that DNA does not depend on mitochondria for it's energy requirements.
- The formation of new chain always takes place in 5'- 3' direction. As a result of this, one chain of DNA is continuously formed and it is termed as leading strand. The formation of second chain begins from the centre and not from the terminal points, so this chain is discontinuous and is made up of small segments called Okazaki fragments. This discontinuous chain is termed as lagging strand. Ultimately, all these segments are joined together and a complete new chain is formed.
Fig. : Continuous replication of a daughter DNA strand on leading strand and discontinuous replication of lagging strand.
- The okazaki segments are joined together by an enzyme DNA ligase (Khorana).
- The formation of new chains is catalyzed by an enzyme DNA Polymerase.
In prokaryotes, it is of 3 types:
- DNA - Polymerase I : This was discovered by Kornberg (1957). This enzyme functions as exonuclease. It separates RNA primer from DNA and also fills the gap. It is also known as DNA-repair enzyme.
- DNA - Polymerase II : It is least reactive in replication process. It is also helpful in DNA-repairing in absence of DNA-polymerase-I and DNA polymerase-III .
- DNA - Polymerase III : This is the main enzyme in DNA-replication. The larger chains are formed by this enzyme. This is also known as replicase. DNA polymerase III is a complex enzyme composed of seven polypeptides α, ε, θ, β, γ1, δ, γ2.
In eukaryotes, there occur five types of DNA polymerase enzyme.
- α-DNA - polymerase = Similar to DNA polymerase I.
- β-DNA - polymerase = It is concerned with DNA repair.
- γ-DNA - polymerase = It is concerned with replication of cytoplasmic DNA.
- δ -DNA - polymerase = It is similar to DNA - polymerase II.
- ε - DNA polymerase = It is similar to DNA - polymerase III.
Thus, DNA replication process is completed with the effect of different enzymes.
In the semi-conservative mode of replication, each daughter DNA molecule receives one chain of polynucleotides from the mother DNA molecule and the second chain is synthesized.
NOTES - All DNA polymerase I, II and III enzymes have 5'-3' polymerisation activity and 3'-5' exonuclease activity.
GENETIC CODE
- The term 'genetic code' was given by George Gamow (1954). He was the first to propose the triplet code (one codon consists of three nitrogen bases).
- The relationship between the sequence of amino acids in a polypeptide chain and nucleotide sequence of DNA or m-RNA is called genetic code.
- There occurs 20 types of amino acids which participate in protein synthesis. DNA contains information for the synthesis of any types of polypeptide chain. In the process of transcription, information transfers from DNA to m-RNA in the form of complementary N2-base sequence.
- A codon is the nucleotide sequence in m-RNA which codes for particular amino acid ; whereas the genetic code is the sequence of nucleotides in m-RNA molecule, which contains information for the synthesis of polypeptide chain.
- Nirenberg gave the first experimental proof for the triplet code.
- 61 out of 64 codons code for only 20 amino acids.
- The main problem of genetic code was to determine the exact number of nucleotides in a codon which codes for one amino acid.
- There are four types of N2-bases in m-RNA (A, U, G, C) for 20 types of amino acids.
Table : Triplet codons of m-RNA for amino acids
CHARACTERISTICS OF GENETIC CODE
TRIPLET IN NATURE
- A codon is composed of three adjacent nitrogen bases which specifies one amino acid in polypeptide chain.
For e.g. : In m-RNA, if there are total 90 N2 - bases. Then this m-RNA determines 30 amino acids in a polypeptide chain.
UNIVERSALITY
- The genetic code is applicable universally.
- The same genetic code is present in all kinds of living organisms including viruses, bacteria, unicellular and multicellular organisms. In all these organisms, triplet code is for specific amino acid.
NON-AMBIGUOUS
- Genetic code is non ambiguous i.e. one codon specifies only one amino acid and not any other.
- In this case, one codon never code two different amino acids. Exception GUG codon which codes both valine and methionine amino acid.
NON-OVERLAPPING
- A nitrogen base is a constituent of only one codon.
COMMA LESS
- There is no punctuation (comma) between the adjacent codon i.e. each codon is immediately followed by the next codon.
- If a nucleotide is deleted or added, the whole genetic code read differently.
- A polypeptide chain having 50 amino acids shall be specialized by a linear sequence of 150 nucleotides. If a nucleotide is added in the middle of this sequence, the first 25 amino acids of polypeptide will be same but next 25 amino acids will be different.
DEGENERACY OF GENETIC CODE
- There are 64 codons for 20 types of amino acids, so most of the amino acids (except two) can be coded by more than one codon. Single amino acid coded by more than one codon is called degeneracy of genetic code.
- Only two amino acids-tryptophan and methionine are specified by a single codon.
- All the other amino acids are specified or coded by 2 to 6 codons.
- Leucine, serine and arginine are coded or specified by
6-codons.
Leucine = CUU, CUC, CUA, CUG, UUA and UUG
Serine = UCU, UCC, UCA, UCG, AGU, AGC
Arginine = CGU, CGC, CGA, CGG, AGA, AGG
- Degeneracy of genetic code is related to third position
(3'-end of triplet codon) of codon. The third base is described as “Wobble base”. - Exception :
- Different codon : Normally UAA and UGA are chain termination codon but in Paramecium and other ciliates, UAA and UGA code for glutamine amino acid.
- Mitochondrial gene : Normally AGG and AGA code for arginine amino acid but in human mitochondria these function as stop codon.
UGA, a termination codon corresponds to tryptophan while AUA (Codon for isoleucine) denotes methionine in human mitochondria.
CHAIN INITIATION AND CHAIN TERMINATION CODON
- Polypeptide chain synthesis is signalled by two initiation codons - AUG or GUG.
- AUG codes methionine amino acid in eukaryotes and in prokaryotes AUG codes N-formyl methionine.
- Sometimes GUG also functions as start codon, it codes for valine amino acid normally but when it is present at starting position it codes for methionine amino acid.
- Out of 64 codons, 3-codons are stopping or nonsense or termination codon.
Nonsense codons do not specify any amino acid.
So only 61 codons are sense codons which specify 20 amino acids.
WOBBLE HYPOTHESIS
- Wobble hypothesis was propounded by Crick (1965).
- Normally an anticodon recognises only one codon, but sometimes an anticodon recognises more than one codon. This is known as wobbling. Wobbling normally occurs for third nucleotide of codon.
- For e.g., anticodon AAG can recognise two anticodons i.e. UUU and UUC, both stand for phenylalanine.
- Nonsense codons lie in middle position in polycistronic m-RNA.
CENTRAL DOGMA
- Central dogma term was given by Crick. It proposes unidirectional or one way flow of information from DNA to RNA and then to protein.
- The formation (production) of m - RNA from DNA and then synthesis of protein from it, is known as the central dogma.
- It means, it includes transcription and translation.
- The central dogma scheme of protein synthesis was presented by Jacob and Monad.
- The detailed study of central dogma is done by Nirenberg, Mathai and Khorana.
- The formation of DNA from RNA is known as reverse transcription. It was discovered by Temin and Baltimore in Rous -sarcoma virus. So, it is also called Teminism.
ss-RNA of Rous-Sarcoma virus (Retrovirus) produces
ds-DNA in host’s cell with the help of enzyme reverse transcriptase (DNA-polymerase). This DNA is called c-DNA (complimentary DNA). Sometimes, this DNA moves in host genome. Such mobile DNA is called retroposon (oncogene).
ds-DNA in host’s cell with the help of enzyme reverse transcriptase (DNA-polymerase). This DNA is called c-DNA (complimentary DNA). Sometimes, this DNA moves in host genome. Such mobile DNA is called retroposon (oncogene).
TRANSCRIPTION
- Formation of RNA over DNA template is called transcription. Out of two strands of DNA, only one strand participates in transcription and called antisense strand.
- Transcription occurs during interphase.
- The segment of DNA involved in transcription is called cistron.
- RNA polymerase enzyme is involved in transcription. In eukaryotes, there are three types of RNA polymerases –
- RNA polymerase-I for 28S rRNA, 18S rRNA, 5.8S rRNA.
- RNA polymerase-II for m-RNA.
- RNA polymerase-III for t-RNA, 5S rRNA. SnRNA
- In eukaryotes, RNA polymerase enzyme is composed of 10-15 polypeptide chains.
- Prokaryotes have only one type of RNA polymerase, which synthesizes all types of RNAs.
- RNA polymerase of E. coli has five polypeptide chains- β, β ', α, α and σ
- σ polypeptide chain is also known as σ factor (sigma factor).
- Core enzyme + sigma factor ⇒ RNA polymerase
(β, β ', α, α) (σ)
STEPS OF TRANSCRIPTION
(1) INITIATION
- DNA has a “promoter site or initiation site” where transcription begins and a “terminator site”, where transcription stops.
- Sigma factor (σ) recognises the promoter site of DNA.
- With the help of sigma factor, RNA polymerase enzyme gets attached to a specific site of DNA called “promoter site”.
- In prokaryotes, before the 10 N2 base from promoter site, a sequence of 6 base pairs (TATAAT) is present on DNA, which is called Pribnow box.
- In eukaryotes, before the 20 N2 base from promoter site a sequence of 7 base pairs (TATAAAA) or (TATATAT) is present on DNA, which is called “TATA box or Hogness box.”
- At promoter site, RNA polymerase enzyme breaks
H-bonds between two DNA strands and separates them.
One of the strand takes part in transcription. Transcription proceeds in 5' → 3' direction.
- Ribonucleotide triphosphate come to lie opposite complementary nitrogen bases of antisense strand.
- These ribonucleotides are present in the form of triphosphate ATP, GTP, UTP and CTP in nucleoplasm. When they used in transcription, pyrophosphatase hydrolyses two phosphates from each activated nucleotide. This releases energy, which is used in the process of transcription.
(2) ELONGATION
- RNA polymerase enzyme establishes phosphodiester bond between adjacent ribonucleotides.
- Sigma factor separates and core enzyme moves along the antisense strand till it reaches the terminator site.
- The elongation requires the bivalent ions Mn++ or Mg2+.
(3) TERMINATION
- When RNA polymerase enzyme reaches terminator site, it separates from DNA template.
- In terminator site on DNA, N2 bases are present in palindromic sequence.
- In most cases, RNA polymerase enzyme can recognise the terminator site and stop the synthesis of RNA chain, but in prokaryotes it recognises the terminator site with the help of Rho factor (ρ factor).
- Rho (ρ) factor is a specific protein which helps RNA polymerase enzyme to recognise the terminator site.
Fig. : Process of Transcription in Bacteria
PROTEIN SYNTHESIS
- Translation (protein synthesis) is the mechanism by which the triplet base sequence of a mRNA guides the linking of a specific sequence of amino acids to form a polypeptide on ribosomes.
- Protein synthesis occurs over ribosomes.
- It consists of ribosomes, amino acids, mRNA, + RNAs and amino acyl tRNA synthetase.
STEPS OF PROTEIN SYNTHESIS
(1) ACTIVATION OF AMINO ACID
- 20 types of amino acids participate in protein synthesis.
- Amino acid reacts with ATP to form amino acyl AMP enzyme complex, which is also known as activated amino acid.
Amino acid + ATP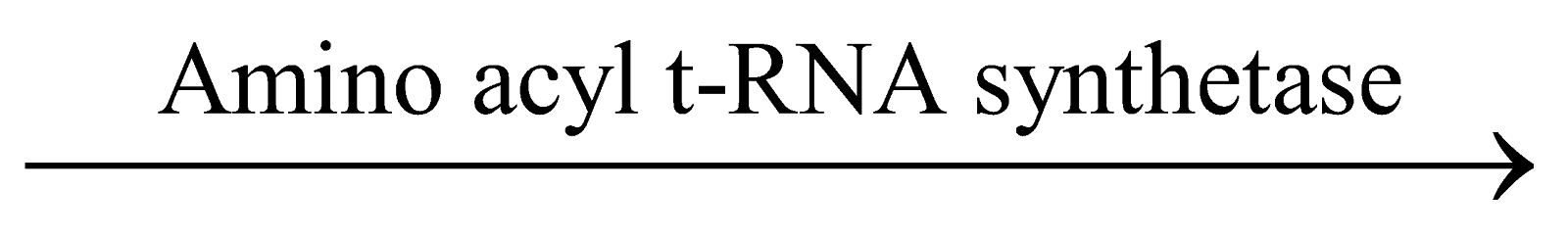 Amino acyl AMP-enzyme complex + PP
Amino acyl AMP-enzyme complex + PP
This reaction is catalyzed by a specific ‘amino acyl
t-RNA synthetase’ enzyme.
t-RNA synthetase’ enzyme.
- There is a separate ‘Amino acyl t-RNA synthetase’ enzyme for each kind of amino acid.
(2) CHARGING OF t-RNA
- Specific activated amino acid is recognised by its specific t-RNA.
- Now amino acid attaches to the ‘amino acid attachment site’ of its specific t-RNA and AMP and enzyme are separated from it.
Amino acyl AMP-enzyme complex + t-RNA ––––→ Amino acyl t-RNA complex + AMP + enzyme
- Amino acyl t-RNA complex is also called charged t-RNA.
Now, Amino acyl t-RNA moves to the ribosome for protein synthesis.
(3) INITIATION OF POLYPEPTIDE CHAIN
- In this step, 30S and 50S subunits of ribosome, GTP, Mg+2, charged t-RNA, m-RNA and some initiation factors are required.
- In prokaryotes, there are three initiation factors :
IF1, IF2 and IF3.
In eukaryotes, there are more than 3 initiation factors.
- GTP and initiation factors promote the initiation process.
- In starting, both subunits of ribosomes are separated with the help of IF3 factor.
- In prokaryotes, with the help of “SD sequence” (Shine-Delgamo sequence), m-RNA recognises the smaller subunit of ribosome. A sequence of 8 nitrogen base is present before the 4-12 nitrogen base of initiation codon on mRNA, called “SD sequence”. In smaller subunit of ribosome, a complementary sequence of “SD sequence” is present on 16S rRNA, which is called anti Shine-dalgarno sequence (ASD sequence).
With the help of SD and ASD sequence, mRNA recognises the smaller subunit of ribosome. While in eukaryotes, smaller subunit of ribosome is recognised by “7mG cap”.
In eukaryotes, 18S rRNA of smaller sub unit has a complementary sequence of “7mG cap”.
30S subunit + m-RNA 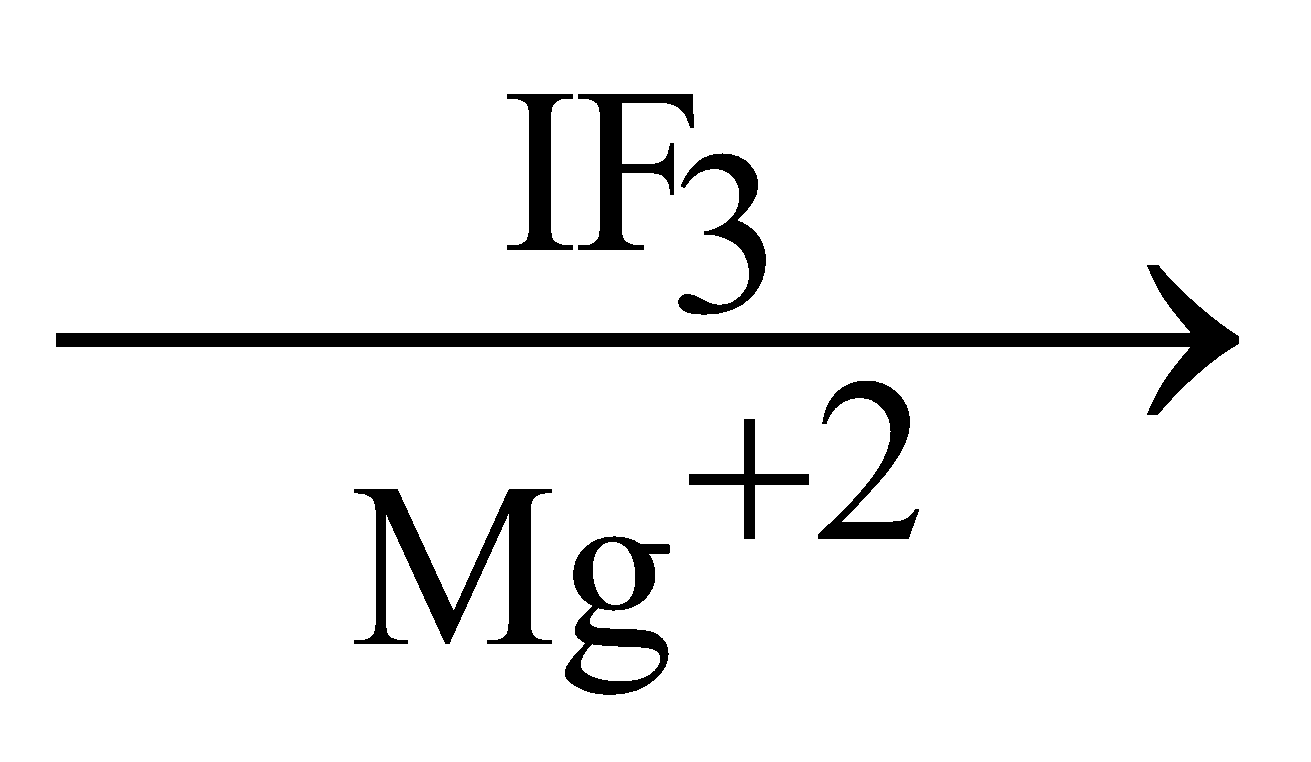 30S m-RNA complex
30S m-RNA complex
This 30 S m-RNA - complex reacts with formyl methionyl t-RNA - complex and 30 S m-RNA - formyl methionyl t-RNA - complex is formed. This t-RNA attaches with codon part of m-RNA. A GTP molecule is required.
30 'S' m-RNA - complex + Formyl methionyl t-RNA-complex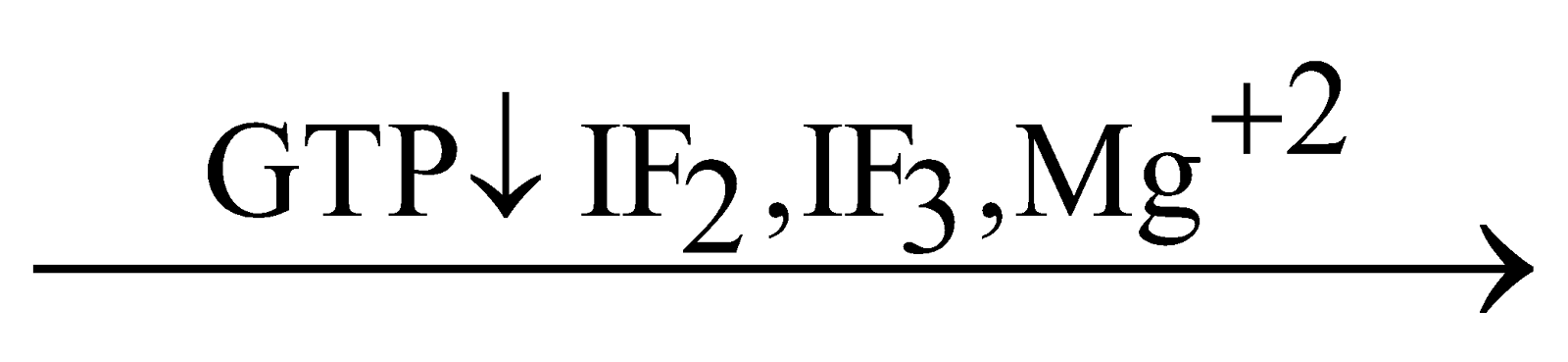 30 'S' m-RNA formyl methionyl t-RNA-complex
30 'S' m-RNA formyl methionyl t-RNA-complex
- Now larger subunit of ribosome (50S subunit) joins this complex. The initiation factor is released and complete 70S ribosome is formed.
- In larger subunit of ribosome, there are three sites for t-RNA -
P site = Peptidyl site.
A site = Aminoacyl site.
E - site = Exit site
- Starting codon of m-RNA is near to P site of the ribosome, so t-RNA with formyl methionine amino acid first attaches to P site of ribosome and next codon of m-RNA is near to A site of the ribosome. So new t-RNA with new amino acid always attaches at A site of ribosome but in the initiation step, A site is empty.
(4) CHAIN-ELONGATION
- Chain elongation requires tRNA, GTP, Mg+2 and elongation factors.
- New tRNA with new amino acid attaches at A site of the ribosome.
- First of all t-RNA of P-site is discharged so – COOH of P-site amino acid becomes free. Now peptide bond takes place between – COOH group of P site amino acid and – NH2 group of A-site amino acid.
- Peptidyl transferase enzyme induces the formation of peptide bond. In peptide bond formation, a 23S r-RNA is also helpful. This r-RNA acts as an enzyme so it is also called ribozyme.
- After formation of peptide bond, t-RNA of P site is released from ribosomes via E-site and dipeptide attaches with A site.
Now t-RNA of A site is transferred to the P site and A site becomes empty.
- Next, ribosome slides over m-RNA strand in 5'-3' direction. Due to sliding of ribosome on m-RNA, new codon of m-RNA is continuously available at A site of ribosome and according to new codon of m-RNA new amino acid attaches in polypeptide chain.
- Translocase enzyme is helpful in movement of ribosome. GTP provides energy for the sliding of ribosome.
- In elongation process, some protein factors are also helpful, which is known as elongation factors.
In prokaryotes, three elongation factors are present:
EF-Tu, EF-Ts, EF-G.
In eukaryotes, two elongation factors are present : eEF1, eEF2.
Fig. : Translation
(5) CHAIN-TERMINATION
- Due to sliding of ribosome over m-RNA, when any nonsense codon (UAA, UAG, UGA) is available at A site of ribosome, then polypeptide chain terminates.
- The linkage between the last t-RNA and the polypeptide chain is broken by three release factor called RF1, RF2, RF3 with the help of GTP.
- Peptidyl transferase enzyme also catalyses the releasing process.
- In eukaryotes, only one release factor is known - eRF1.
- Once the initial portion of mRNA is translated, it is free to engage into another round of translation. This process can be repeated several times, creating a structure called polyribosomes or polysomes.
ONE GENE ONE ENZYME HYPOTHESIS
- The concept that genes have the information to produce enzymes, or gene metabolism relationship was experimentally proved by Beadle and Tatum (1948), on the basis of experiments conducted on pink bread mould (Neurospora crassa).
- This mould can normally grow in a simple minimal medium containing salts and sugar, making all other chemicals such as amino acids, purines, pyrimidines etc. through enzyme catalysed reactions. This wild type of the mould is called prototroph.
- Beadle and Tatum exposed the pink bread mould to X-rays, which can bring about a change in the nucleotide sequence of DNA and thus, causes mutation. They found that the mutants were unable to grow on a minimal medium. Each type of mutant required some extra nutrients in the minimal medium for its normal growth. Such nutritional mutants are called auxotrophs. They obtained different nutritional mutants requiring amino acids ornithine or citrulline or arginine for growth.
The mutants could be classified into three types
- Mutant I : Some could grow on ornithine, or citrulline, or arginine- containing medium,
- Mutant II : Some could grow on citrulline or arginine containing medium, and
- Mutant III : Some could grow only on arginine supplemented medium.
BIOCHEMICAL PATHWAY
- This shows that all mutants could grow on arginine supplemented medium suggesting that arginine is the final product of this pathway.
- The mutant of class I, could grow on ornithine, citrulline or arginine, suggesting that they lacked the capacity to synthesize ornithine, but beyond that they could complete the pathway.
- Similarly, mutants of class II, could not convert ornithine to citrulline, but if supplied, citrulline could synthesize arginine. The last mutants of class III could not convert citrulline to arginine and had to be supplied latter for growth.
- Beadle and Tatum reasoned that these defects could arise due to defective enzymes in each case. Since such changes were mutational, they held that one gene controls one enzyme in a pathway leading to their famous ‘one gene one enzyme hypothesis’.
TYPES OF GENE
- All the genes do not play the same role nor all genes are active all the time.
- With regard to their role and activity, the genes are of following types:
- Homeotic genes : Homeotic gene regulates the organ differentiation in embryo. Homeobox is related to transcription of homeotic gene.
- Constitutive genes (House-keeping genes) : These genes are expressed constantly, because their products are constant needed for cellular activity. E.g. genes for glycolysis, gene of ATPase enzyme.
- Non-constitutive genes (Smart gene or Luxury gene): These genes remain silent and are expressed only when the gene product is needed. They are switched 'on' or 'off' according to the requirement of cellular activities. Non-constitutive genes are of two types - inducible and repressible.
The inducible genes are switched on in presence of a chemical substance called inducer, required for the functioning of gene activity. E.g., gene of lac operon. The repressible genes continue to express themselves till a chemical, often an end product of the metabolism inhibits or represses their activity. E.g., gene of tryptophan operon.
- Overlapping gene : A few genes in certain bacteria and animal viruses code for two different polypeptides. These are called overlapping genes. For example – φ × 174 virus, SV-40 virus.
- Pseudoallele : Gene which is located on non homologous chromosome or gene which is located on different locus on homologous chromosome produces almost same phenotype called as pseudoallele. Pseudo allele is non-allelic gene that produces identical phenotype in cis-trans position. E.g., Duplicate gene.
- Isoallele : If several alleles exhibit same phenotype then they are said to be is oalleles. For e.g., In Drosophila allele - W+C, W+S, W+g → produce red eye colour.
- Hybrid vigour/Heterosis : Superiority of offsprings over its parents is called as hybrid vigour and it develops due to heterozygosity.
Hybrid vigour can be maintained for a long time in vegetatively propagated crops.
Hybrid vigour can be lost by inbreeding (selfing) because inbreeding induces the homozygosity in offspring.
- Jumping genes : It is a segment of DNA which moves from one chromosome to another chromosome within the genome of an individual. McClintock (1983) got nobel prize for the discovery of jumping gene in maize.
GENE EXPRESSION AND REGULATION
- Gene expression is the mechanism at the molecular level by which a gene is able to express itself in the phenotype of an organism.
- The mechanism of gene expression involves biochemical molecules.
- Different genes in an organism are meant for the synthesis of different proteins.
- It consists of synthesis of specific RNAs, polypeptide, structural proteins, proteinaceous biochemical, enzymes which control the structure or functioning of specific traits.
Fig. : Mechanisms of gene expression
- A variety of mechanisms are now known which regulate gene expression at different levels including transcriptional processing of mRNA and translation.
- The regulation of gene expression in bacteria is called operon system.
OPERON CONCEPT
- Operon concept was proposed by Jacob and Monad in E.coli.
- Segment of genetic material that acts as a regulated unit is called operon.
- The mechanism of regulation of protein synthesis utilizing operon model can be illustrated using two examples (lac and tryptophan) in bacteria.
- Operon are of two types- inducible operon and repressible operon.
INDUCIBLE OPERON
- Inducible operon system regulate genetic material which remains switched off normally but becomes operational in the presence of inducer. E.g., Lac operon of E. coli.
- Lac operon of E. coli have four components-
- Regulatory gene : It synthesizes the repressor that attach with the operator gene and block the passage of RNA polymerase.
- Promotor gene : It provides site for attachment to RNA polymerase enzyme and form the initiating point for the transcription.
- Operator gene : It controls the activities of structure gene and provides the passage to the RNA polymerase enzyme.
- Structural gene : It have three structural genes cistron-Z, cistron - Y, cistron -X.
Cistron-Z is responsible for the synthesis of β galactocidase enzyme.
Cistron-Y is responsible for synthesis of permease enzyme.
Cistron-X is responsible for the synthesis of transacetylase enzyme.
- This lac operon normally remains inactive when lac operon contacts with lactose. The lactose act as an inducer. Lactose combines with the repressor so repressor is detached from operator gene and RNA polymerase enzyme gets its passage and reachs to the structural genes and transcription begins.
- Structural gene form the polycistronic mRNA and form the β-galactocidase enzyme, permease and transacetylase enzyme.
β-galatocidase enzyme hydrolyses lactose into glucose and galactose. Permease enzyme increases the entry of lactose inside the bacterial cell. Transacetylase enzyme has no role in lactose hydrolysis.
Fig. : The lac operon
REPRESSIBLE OPERON
- Operon generally remains active and synthesizes product, but when product crosses the threshold value, operon becomes inactive. E.g., tryptophan operon (Trp. operon)
- Trp. operon consists of four components :-
- Regulatory gene : It synthesizes the aporepressor. Aporepressor can not combine with the operator gene and does not block the passage of RNA polymerase enzyme.
- Promotor gene : It is the initial point of transcription and provides site for attachment to RNA polymerase enzyme.
- Operator gene : It controls the activity of structural genes and provides the passage to the RNA polymerase enzyme.
- Structural gene : Structural gene of Trp. operon are of five types- cistron - E, D, C, B, A.
In Trp. operon, passage of RNA polymerase enzyme generally opens and transcription continuously occurs. All the cistron of Trp. operon form polycistronic mRNA which synthesize tryptophan.
When tryptophan cross threshold value, then it acts as a corepressor. Corepressor combines with the aporepressor and forms the repressor.
Repressor attached with the operator gene blocks the passage of RNA polymerase enzyme and transcription stops thus, operon becomes inactive.
Table : Differences between induction and repression
GENE EXPRESSION IN EUKARYOTES
- The genome of higher eukaryotes is very complex.
- In eukaryotes, functionally related genes may not be clustered together constituting an operon.
- The most popular model is known as ‘Britten-Davidson model’ or ‘Gene battery model’ proposed by Britten and Davidson in 1969.
- A set of structural genes controlled by one sensor site is termed as battery.
- Gene-battery model assumes the presence of four classes of sequences-
- Producer gene: A producer gene is comparable to structural gene of prokaryotic operon.
- Receptor site: A receptor site is comparable to operator gene of bacterial operon and one such receptor site is assumed to be present adjacent to each producer gene.
- Integrator gene: Integrator gene is comparable to regulator gene and is responsible for synthesis of an activator RNA. It activates the receptor site.
- Sensor site: A sensor site regulates the activity of integrator gene. Activator gene can be transcribed only when the sensor site is activated.
The sensor sites are recognized by agents which change the patterns of gene expression like hormones and proteins. When a transcription factor (protein, hormone) binds to the sensor site it causes the transcription of an integrator.
HUMAN GENOME PROJECT
- Human Genome Project (HGP) was an international collaborative research program whose goal was to complete the mapping and understanding of all the genes of human beings. All our genes together are known as genome.
- Genetic make-up of an organism or an individual lies in the DNA sequences. If two individuals differ, then their DNA sequences should also be different, at least at some places. These assumptions led to the quest of finding out the complete DNA sequence of the human genome.
With the establishment of genetic engineering techniques where it was possible to isolate and clone any piece of DNA and availability of simple and fast techniques for determining DNA sequences, a very ambitious project of sequencing the human genome was launched in the year 1990.
- The sequencing of the 3 billion bases of DNA in the human genome will allow researchers to identify human genes, assisting our understanding of the genetic basis of disease and of the key biochemical and developmental processes of the human body.
- HGP was closely associated with the rapid development of a new area in biology called bioinformatics.
- Bioinformatics is a broad term to describe applications of computer technology and information science to organize, interpret and predict biological structure and function. It is usually applied in the context of analyzing DNA sequencing date.
- Some of the important goals of HGP are -
- Identify all the genes (approx. 20000-25000) in human DNA.
- Determine the sequences of the 3 billion chemical base pairs that make up human DNA.
- Store information in databases.
- Improve tools for data analysis.
- Transfer related technologies to other sectors, such as industries.
- Address the ethical, legal, and social issues (ELSI) that may arise from the project.
- Many non-human model organisms, such as bacteria, yeast, Caenorhabditis elegans (a free living non-pathogenic nematode), Drosophila (the fruit fly), plants (rice and Arabidopsis), etc., have also been sequenced.
- The method HGP involved two major approaches-
- Expressed Sequence Tags (ESTs): It involves identifying all the genes that are expressed as RNA.
- Sequence Annotation : It involves the sequencing of the whole set of genome that contained all the coding and non-coding sequence, and later assigning different regions in the sequence with functions. For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes and cloned in suitable host using specialised vectors. The cloning resulted into amplification of each piece of DNA fragment so, that subsequently could be sequenced with ease. The commonly used hosts were bacteria and yeast, and the vectors were called BAC (bacterial artificial chromosomes), and YAC (yeast artificial chromosomes).
The fragments were sequenced using automated DNA sequencers that worked on the principle of a method developed by Frederick Sanger. These sequences were then arranged based on some overlapping regions present in them. This required generation of overlapping fragments for sequencing and for aligning these sequences specialised computer based programmes were developed. These sequences were subsequently annotated and were assigned to each chromosome. The sequence of chromosome I was completed only in May 2006 (this was the last of the 24 human chromosomes -22 autosomes and X and Y to be sequenced). Another challenging task was assigning the genetic and physical maps on the genome. This was generated using information on polymorphism of restriction endonuclease recognition sites, and some repetitive DNA sequences known as microsatellites.
Fig. : A representative diagram of human genome project
SALIENT FEATURES OF HUMAN GENOME
- The human genome contains 3164.7 million nucleotide bases.
- The average gene consists of 3000 bases, but sizes vary greatly, with the largest known human gene being dystrophin at 2.4 million bases.
- The total number of genes is estimated at 30,000-much lower than previous estimates of 80,000 to 1,40,000 genes. Almost all (99.9 percent) nucleotide bases are exactly the same in all people.
- Less than 2 percent of the genome codes for proteins.
- Repeated sequences make up a very large portion of the human genome. Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times. They are thought to have no direct coding functions, but they shed light on chromosome structure, dynamics and evolution.
- Chromosome 1 has the most genes (2968) and the Y has the fewest (231).
- Scientists have identified about 1.4 million locations where single-base DNA differences (SNPs- single nucleotide polymorphism, pronounced as 'snips') occur in humans. This information promises to revolutionise the processes of finding chromosomal locations for disease-associated sequences and tracing human history.
APPLICATION OF HGP
- Identification of genes that confer susceptibility to certain diseases and enable 80 individuals to take preventive measures.
- Identification of defective genes and hence the opportunity to offer early treatment.
- Prediction of proteins that the genes produce giving an opportunity of designing appropriate drugs to enhance or inhibit the activities of these proteins.
- First prokaryotes in which complete genome was sequenced is Haemophilus influenzae.
- First eukaryote in which complete genome was sequenced is Saccharomyces cervisiae (yeast). ,
- First plant in which complete genome was sequenced is Arabidopsis thaliana (small mustard plant).
- First animal in which complete genome was sequenced is Caenorhabditis elegans (nematode).
- β-globin and insulin gene are less than 10 kilo base pair. T.D.F. gene is the smallest gene (14 base pair) and Duchenne muscular dystrophy gene is made up of 2400 kilo base pair (longest gene).
GENE LIBRARY (GENOMIC DNA LIBRARY)
- A gene library is a collection of many of the desired genes of DNA fragments maintained in clones of bacterial or some other cells.
- It is prepared by the following method :-
DNA fragments containing one or few desired genes are obtained with the help of specific restriction endonucleases. Each fragment is joined to a suitable vehicle DNA to form recombinant DNAs of different nature. These are then introduced into host (bacterial, yeast, plant or animal) cells. The cells containing recombinant DNAs are allowed to multiply in cultures. This will produce clones of cells where the daughter cells carry the same genes which are identical to those of parent cells. A collection of clones with recombinant DNA containing desired genes is a gene library.
- Gene banks is a store house of clones of known DNA fragments, genes, gene maps, seeds, spores, frozen sperm or eggs or embryos. These are stored for possible use in genetic engineering and breeding experiments where species have become extinct. The need of gene banks is being increasingly felt as the rate of extinction is increasing day by day. The human genome project is the most remarkable contribution in this field.
DNA FINGERPRINTING / DNA TYPING / DNA PROFILING/ DNA TEST
- DNA fingerprinting is a technique to identify a person on the basis of his/her DNA specificity.
- This technique was invented by Sir Alec. Jeffery (1984).
- In India, DNA fingerprinting has been started by Dr. V.K. Kashyap and Dr. Lal Ji Singh.
- DNA of humans is almost the same for all individuals but very small amount that differs from person to person that forensic scientists analyze to identify people.
These differences are called polymorphism (many forms) and are the key of DNA typing. Polymorphisms are most useful to forensic scientist. It consists of variation in the length of DNA at specific loci and is called restricted fragment. It is the most important segment for DNA test made up of short repetitive nucleotide sequences, these are called VNTRs (variable number of tandem repeat).
- VNTR's, also called minisatellites, were discovered by Alec Jeffery. Restricted fragment consists of hypervariable repeat region of DNA having a basic repeat sequence of 11-60 bp and flanked on both sites by restriction site. The number and position of minisatellites or VNTR in restriction fragment is different for each DNA and length of restricted fragment depend on number of VNTR.
Therefore, when the genome of two people are cut using the same restriction enzymes, the length of fragments obtained is different for both the people.
Fig. : Schematic representation of DNA fingerprinting: Few representative chromosomes have been shown to contain different copy number of VNTR. For the sake of understanding, different colour schemes have been used to trace the origin of each band in the gel. The two alleles (paternal and maternal) of a chromosome also contain different copy numbers of VNTR. It is clear that the banding pattern of DNA from crime scene matches with individual B, and not with A.







